I have supervised various undergraduate students' research training projects, including statistical and computational methodologies on modeling zero-inflated data, multivariate regression, low-rank approximation, genetic association studies, etc. I am committed to learning what each student needs to succeed and encouraging active learning skills to carry forward. Here are some examples. More on the way!
Research opportunities are open to highly motivated students. Interested individuals are encouraged to reach out for more details.
Bear food use regression analysis (STAT 341 Honors Option)
-
Mentored student: Danielle Terry (CSU undergraduate, Fall 2025)
-
Summary: Using camera trap detections and paired berry surveys collected in the Methow Valley (May–Oct 2024), this project explored how bear detections relate to environmental and human factors. The student conducted exploratory analyses and correlation screening, then compared a multiple linear regression model for monthly detection counts with a Poisson generalized linear mixed model (effort offset and site-level random intercept). The analysis highlights consistent associations of bear detections with proximity to development and serviceberry abundance, and a pronounced seasonal pattern (July vs June).
-
Note: Project report (PDF)
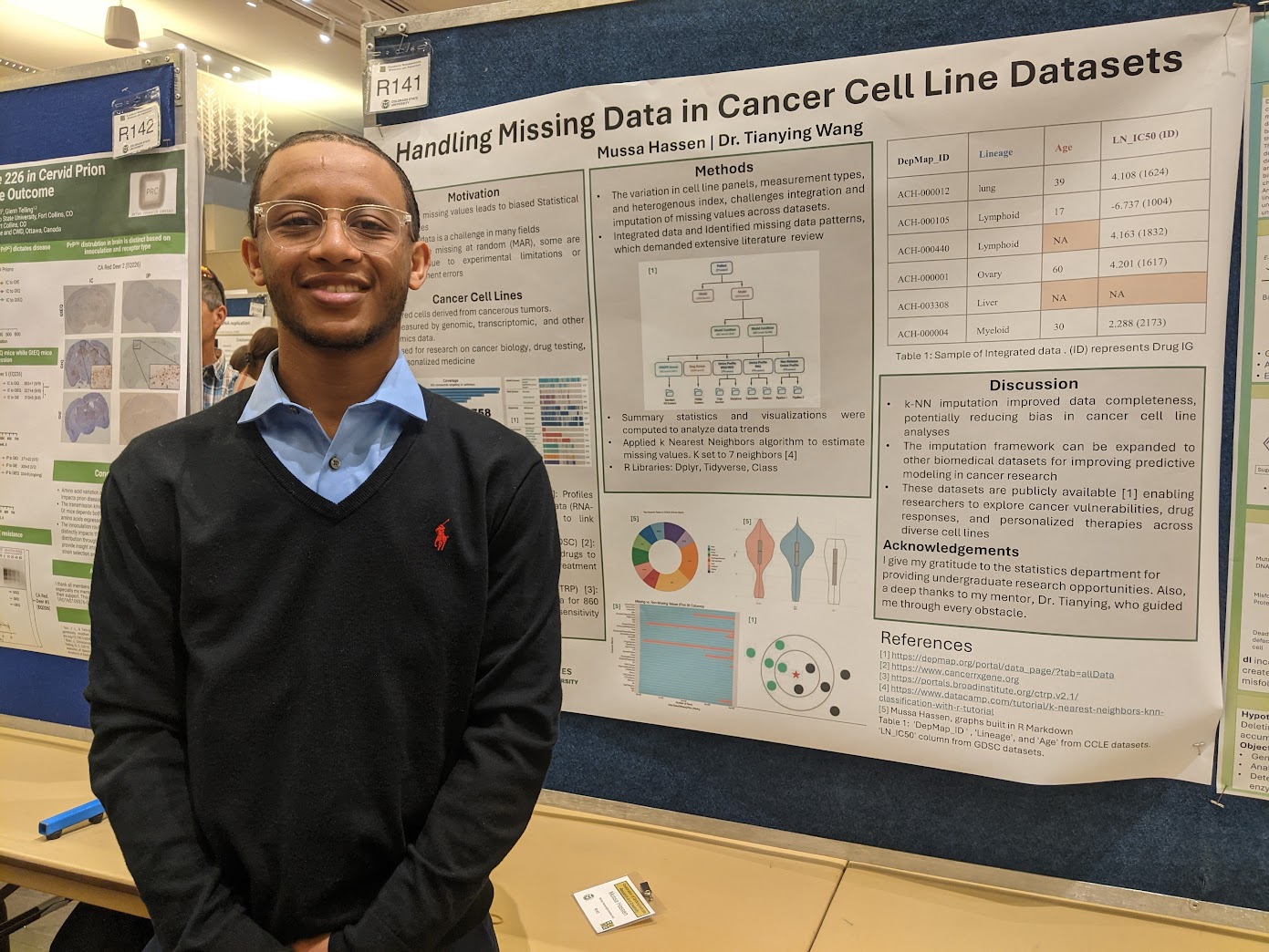
Handling Missing Data in Cancer Cell Line Datasets
-
Mentored student: Mussa Hassen (2024 Summer, undergraduate students’ research program at CSU)
-
Summary: Large, publicly available cancer-cell-line resources fuel precision-oncology discovery yet suffer from pervasive, non-ignorable missing values that distort downstream analyses of multi-omics profiles and drug responses. In this project, we harmonized these heterogeneous panels, mapped their complex missing-data mechanisms, and benchmarked a comprehensive suite of statistical and machine-learning imputation schemes. The resulting pipeline, highlighted in a CURC poster, offers a transferable template for rigorously handling missingness in other large-scale biomedical datasets.
-
Note: Mussa won the 2025 Undergraduate Excellence in Research Award for his efforts in this project. Congratulations, Mussa!
 Photo courtesy of CSU Department of Statistics — 2025 Statistics Awards Ceremony
Photo courtesy of CSU Department of Statistics — 2025 Statistics Awards Ceremony
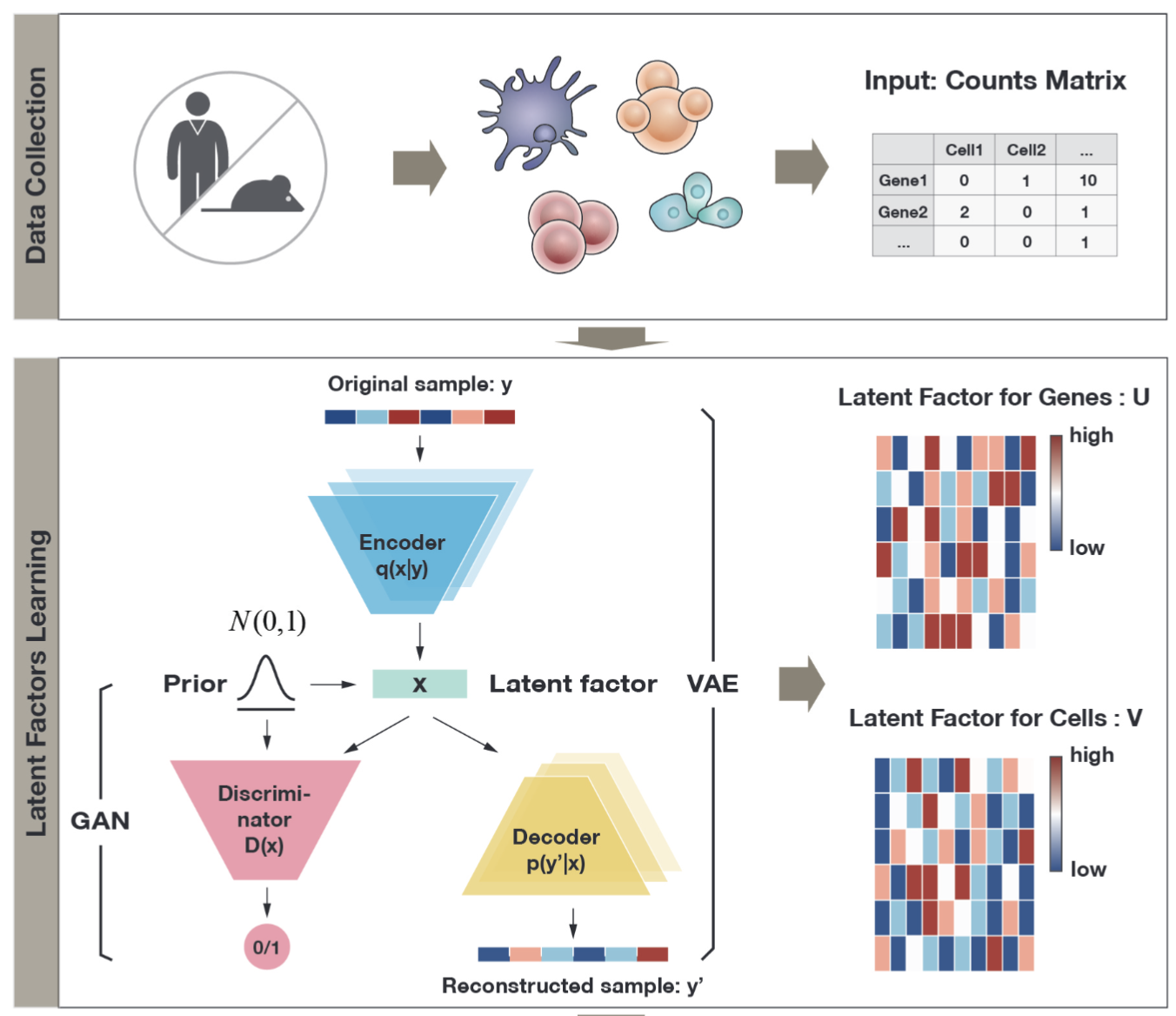
Deep learning-based statistical modeling for data denoising and imputation
- Mentored students: Qinhuan Luo and Yongzhe Yu. (This work was finished when Qinhuan and Yongzhen were undergraduate students at Tsinghua University from 2022-2023.)
-
Summary: Single-cell RNA sequencing data is a vital and rich resource for researchers to investigate the underlying mechanisms of various biological or disease-related processes. However, the well-known data heterogeneity is an unignorable barrier and causes many challenges in related studies. The heterogeneity can arise from multiple sources, such as sequencing techniques and platforms, and is reflected in cell-wise or gene-wise heterogeneity. To mitigate the modeling limitations caused by unobserved confounders and unexplored transcriptomic pathways, we propose a novel framework to model the complex heterogeneity in scRNA-seq data by integrating deep generative models with parametric statistical models. By comprehensively evaluating the proposed method on four datasets, we demonstrated its superior ability to model different sources of data heterogeneity at different levels of known information.
-
Related paper: Luo, Q.♦, Yu, Y.♦, and Wang, T.✉ (2025). “Denoising Single-Cell RNA-Seq Data with a Deep Learning-Embedded Statistical Framework”, BMC Bioinformatics, accepted.
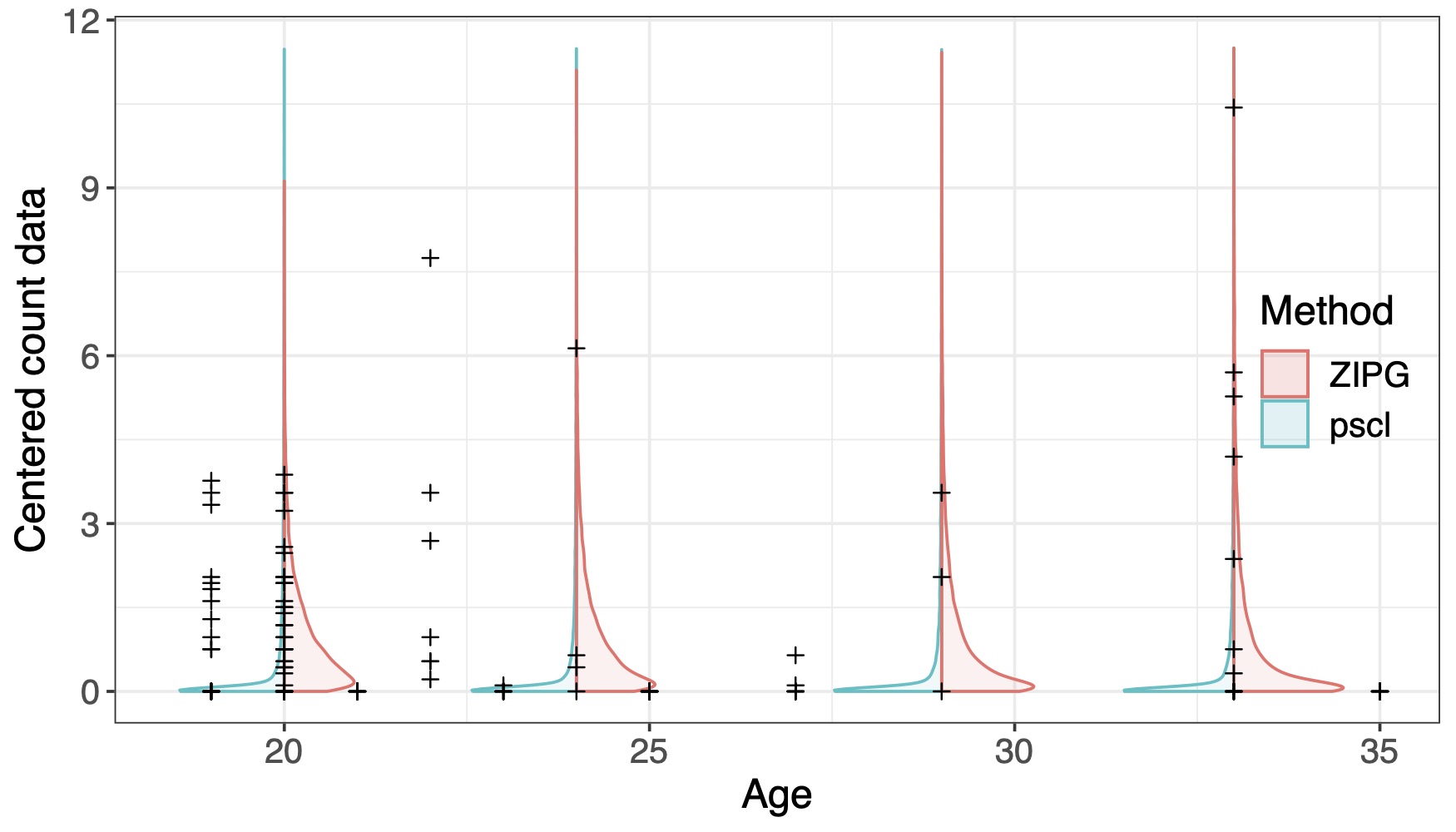
Modeling zero-inflated data with error-in-variables
- Mentored student: Roulan Jiang. (This work was conducted when Roulan was a senior undergraduate student at Tsinghua University from 2021-2022.)
-
Summary: In microbiome studies, it is of interest to use a sample from a population of microbes, such as the gut microbiota community, to estimate the population proportion of these taxa. However, due to biases introduced in sampling and preprocessing steps, these observed taxa abundances may not reflect true taxa abundance patterns in the ecosystem. Repeated measures, including longitudinal study designs, may be potential solutions to mitigate the discrepancy between observed abundances and true underlying abundances. Yet, widely observed zero-inflation and over-dispersion issues can distort downstream statistical analyses aiming to associate taxa abundances with covariates of interest. We propose a Zero-Inflated Poisson Gamma (ZIPG) framework to address the aforementioned challenges. The above figure shows how the estimated differential variability of a taxon in pregnant women changes over age.
-
Related paper: Jiang, R.♦, Zhan, X.✉, and Wang, T.✉ (2023). “A Flexible Zero-Inflated Poisson-Gamma Model with Application to Microbiome Read Count Data”, Journal of the American Statistical Association, 118 (542), 792 - 804.
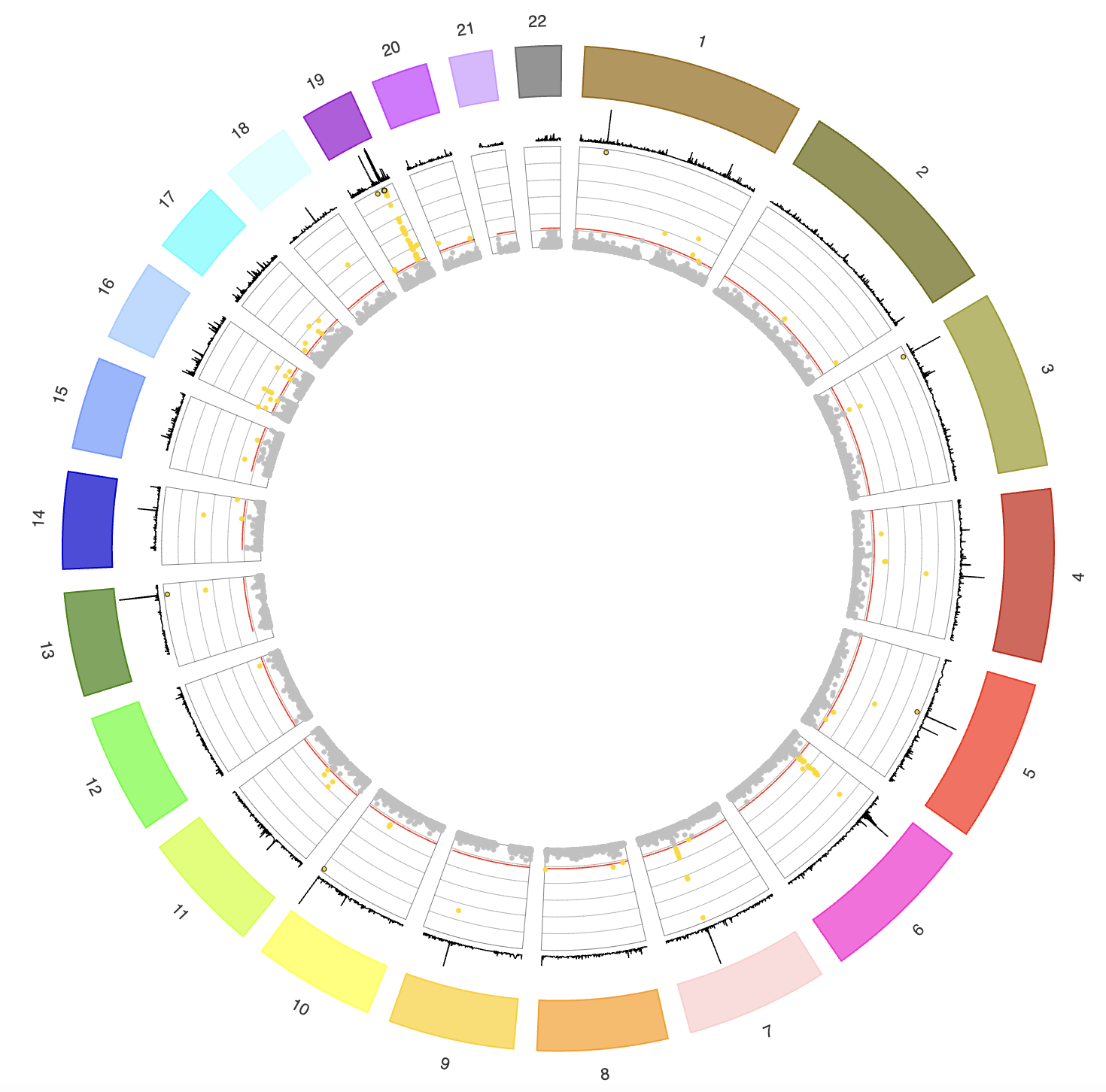
Quantile TWAS Genome Browser (Shiny app)
- Mentored student: Weijing Yin. (This work was conducted as a summer project in 2022 when Weijing was a sophomore undergraduate student at Tsinghua University.)
-
Summary: This visualization is based on QTWAS, a quantile regression model for Transcriptome-wide Association Studies (TWAS). Here we provide QTWAS results on the summary statistics from ten GWAS studies on brain disorders, including five neuropsychiatric traits: schizophrenia (SCZ), attention-deficit/hyperactivity disorder (ADHD), bipolar disorder (BD), autism spectrum disorder (ASD), and major depressive disorder (MDD); and four neurodegenerative traits: Alzheimer’s disease (AD_Kunkle and AD_Jansen), Parkinson’s disease (PD), multiple sclerosis (MS) and amyotrophic lateral sclerosis (ALS).
-
App link: tianyingw.shinyapps.io/QTWAS/.
Estimating quantile curves for zero-inflated outcomes
- Mentored student: Zirui Wang. (This work was finished when Zirui was a junior undergraduate student at Tsinghua University from 2021-2022.)

-
Summary: We consider the complex data modeling problem motivated by the zero-inflated and overdispersed microbiome read count data. Several parametric approaches have been proposed to address issues of zero inflation and overdispersion, such as zero-inflated Poisson regression and zero-inflated Negative Binomial regression. However, parametric assumptions could be easily violated in real-world applications. In this project, we propose a semiparametric single-index quantile regression framework, which is flexible in including a wide range of possible association functions and adaptable to the various zero proportions across subjects. We establish the asymptotic normality of the index coefficients estimator and the asymptotic convergence rate of the nonparametric quantile regression curve estimation. Through Monte Carlo simulation studies and the application in a microbiome study, we demonstrate the superior performance of the proposed method. The above figure shows how our proposed method fits the real microbiome data (Roseburia) better than other methods.
-
Related paper: Wang, Z.♦ and Wang, T.✉ (2024). “A Semiparametric Quantile Single-Index Model for Zero-Inflated Outcomes”, Statistica Sinica, accepted.
-
Notes: Zirui won the Best Student Paper Award at the 2024 National Graduate Statistics Symposium, Mathematical Statistics Section, based on this work. Congratulations, Zirui!